“TREC” Description
1.1 Overall objective of the project
Traversing European Coastlines (TREC) is an international and highly collaborative scientific expedition and multidisciplinary scientific project to study life in coastal ecosystems.
Coastal areas are under direct threat by human activity, such as pollution, agriculture and urbanisation, and are strongly affected by global impacts such as the effects of climate change. Despite decades of excellent research into our oceans and soils at coastal regions, there are still many unanswered but important questions, especially about the impact of human activity on biodiversity at microbial scale and about the molecular mechanisms involved in organismal-environment feedback. The TREC project has a focus on understanding coastal biodiversity and its response to natural and man-made environmental factors, from molecules to ecosystems across the European coasts.
In a nutshell TREC:
- Studies life in its natural context at various scales and links it to the physical and chemical environment.
- Brings molecular sciences to environmental research in an unprecedented way.
- Quantifies the impact of natural and human-made factors on life in the two main ecosystems on earth, land and sea.
- Explores the interface of land and sea to address how activities on land affect the sea ecosystems and vice versa, at microscopic and molecular level.
- Implements an open, holistic and systemic approach at unprecedented (continental) scale to capture the interconnectivity of many local and global processes.
- Reaches out to citizens and policymakers to explain the coupling of human and planetary health.
The EMBL has gathered, after four years of intense preparation phase, the tools, knowledge, the protocols, logistics experience, technologies and European network (including 2 local partners in Greece) to document, examine, and probe the molecular state of the coastal areas and assess biodiversity.
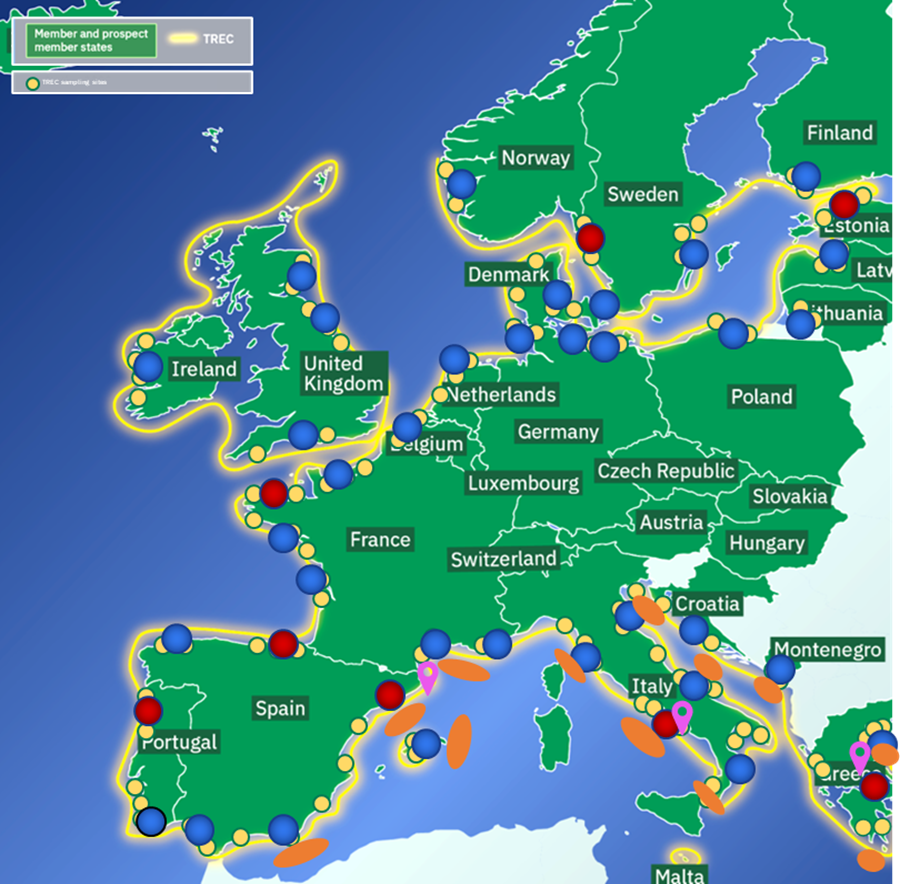
Using a fleet of customised EMBL mobile labs and vehicles and the Tara schooner, researchers will collect water, sediments, soil and air samples, selected species and broad environmental data from coastal land areas, shorelines, inland and offshore waters, along the coast of Europe. The sampling involves highly standardised cross-sectional sampling and protocols to cover all organismal scales from viruses to animals, plants and macroalgae. TREC sampling started in March 2023 and will end in July 2024 by sampling at around 120 sites in 22 European countries, involving over 200 scientists in the field, a team of experts to run cutting-edge technologies and an international science education and public engagement programme.
All samples collected during TREC will be used for fundamental and non-commercial research purposes. Analysis will include metagenomics, metabarcoding, metatranscriptomics, metabolomics and imaging analysis (by means of light and electron microscopy). Samples will be also used for bio-geochemical analysis (such as dissolved and particulate organic matter and nutrients) and chemical profiling (including organic and inorganic pollutants, with targeted and untargeted analysis). Environmental data will be taken and metadata will be recorded.
TREC will generate open datasets describing the pretty much unknown microbial biodiversity, but also including macro taxa biodiversity in combination with environmental and metadata collection. All datasets and discoveries will be of open access to enrich the discovery of coastal biodiversity to help finding indicators of coastal ecosystem health and mitigation strategiesr

1.2 Research goals of TREC
- Identification of key factors (natural and man-made) driving coastal community composition along land sea gradients
- Tracking gene flux between soil and ocean species via horizontal gene transfer or species assimilation, both regionally and globally
- A better definition of coastal ecosystems biodiversity, causes of biodiversity loss, and its impact
- Understanding molecular mechanisms of phenotypic plasticity and adaptation at species and community levels
- Elucidating mechanisms of co-adaptation across species and communities
- Unravelling new molecular mechanisms from coastal species and symbionts
- Providing open access to research results, issue scientific publications in scientific journals and promote further research activities
1.3 Researched taxa
TREC aims to research the following taxa (the followings are targeted by TREC in general, for exact sampling details relevant to the location subject of this document, please refer to 4.1 Sampling locations in this document.
TREC covers all organismal scales from viruses, bacteria, archaea, fungi, protists (unicellular eukaryotes) to multicellular eukaryotes.
Planktonic organisms that will be collected:
- Dinoflagellates
- Ciliates
- Picoeukaryotes
- Haptophytes
- Acantharians
- Diatoms
- Ichtyosporans
Multicellular organisms that will be collected in marine habitats:
- Sponges
- Annelids (with focus on Platynereis dumerilii)
- Nematodes
- Sea grasses (such as Zostera marina, Posidonia oceanica)
- Diverse macroalgae
- Brown algae
- Lucinid clams
- Some cnidarian species (such as Anemonia viridis)
- Lichens (such as Trebouxia and Physcia)
Live species will be also collected for culturing and further analysis including:
- Viruses of unicellular eukaryotes
- Bacteria
- Alveolates such as Uronema marinum, Miamiensis avidus, and Perkinsus sp, phytoplankton
- Marine (non-metazoan) holozoans such as Sphaeroforma arctica and Chromosphaera perkinsii
- Platynereis dumerilii
- Planarians
- Fungi
Leaves of non-protected terrestrial plants will also be collected, species depend on availability. Soundscape will be also recorded by non-invasive methods and analysed covering all sounds in the environment both in marine and terrestrial habitats.
None of the taxa are endangered/protected as defined by the Washington Convention (also known as CITES), nor are endangered/protected species intended to be sampled.
2. Sampling plans, downstream processing and analysis
The expedition will combine ocean exploration, conducted by the Tara Ocean Foundation (Tara Europa expedition), with parallel land sampling of soil, sediment, shallow waters, aerosols and selected species in various habitats.
Broad environmental data collection by ground sensors and satellite measurements. Sampling is highly standardised to enable comparability and integration of samples at continental scale.
(The application for research authorization for the Tara Europa sampling component is being handled by the Tara Ocean Foundation and being sent to the relevant authorities).
All samples will be processed and preserved in different conditions for a variety of downstream analysis including:
- Environmental Metagenomes done by deep sequencing with short reads, long reads and HiC may be performed in some samples. The main focus is on microbial biodiversity and community composition.
- Metabarcoding by deep sequencing with primers for 16S, 18S and ITS full length seq
- Environmental Metatranscriptomes with enrichment in prokaryotes and eukaryotes
- Environmental Metabolomics by mass spec
- Single cell sequencing of selected species (for single cell organisms ecology and single cell adaptations in response to chemical pollution and other environmental factors)
- Single individuals low genome sequencing (eg for population genetics and biogeography)
- Expansion microscopy of plankton
- Light and electron microscopy imaging for taxonomic and phenotypic purposes, from subcellular ultrastructures to phenotypes at individual or population level.
- Live organisms culturing
- Biogeochemical analysis (such as nutrients, DOC, CDOM, FDOM, pyrogenic carbon)
- Chemical profiling including non-targeted and targeted mass spec analysis. Targeted analysis includes a unique library of more than 1000 environmental compounds including pesticides or non-degradable pharmaceuticals/antibiotics and their degradation products to targeted analysis
TREC allows novel combinations of various cutting edge technologies, normally not available at the sampling sites, to study microbes and selected species with novel approaches to associate morphological with genomic features as well as taxonomy with functional traits. The analyses are done alone the land sea gradients at each sampling site and along broader environmental gradients along the European coastline providing unique correlations between morphological and genomics features at all organismal scales with various environmental data at continental scale. This study focuses more specifically on the mechanisms of antibiotics resistance spread across different ecosystems and compare with their acquisition by human microbes (e.g. from farm to fork) and with human-caused antibiotic resistance spread (e.g. through waste water) and allows to characterise and connect microbes with macroscopic taxa to better understand their roles in evolution, assimilation and behaviour.
2.1 Environmental samples / land-sea interface sampling
2.1.1 Sampling soil, superficial intertidal and/or subtidal sediments
Coastal regions represent a dramatic natural habitat transition to which organisms have adapted; regions that have been more recently intensively affected by human activities. Anthropogenic pollutants are spread throughout the coastal environment through groundwater, watersheds, rivers, and accumulate in soil and sediments where they can alter the composition and species abundances within microbial communities. Studying the extent of this effect is often the result of the interplay of several factors including the history of anthropogenic pollution in relation to urbanisation and industrialisation of the coasts, the geomorphological and hydrological factors of the land and water interface and an incomplete understanding of the cause-effect relationship between pollution and microbial community composition in the environment.
For this reason, our study attempts to perform a large-scale analysis of diverse human-impacted and pristine environments across the European coasts in 120 sites, in order to better understand the complex relationships between the soil, sediments and the oceanic microbial communities in diverse coastal environments. We will analyse the structuration of microbial communities in superficial sediments and soils in relation to human impacts and pollution, seeking for a general biodiversity and pollutant pattern across Europe.
Methods of soil and sediment sample collection
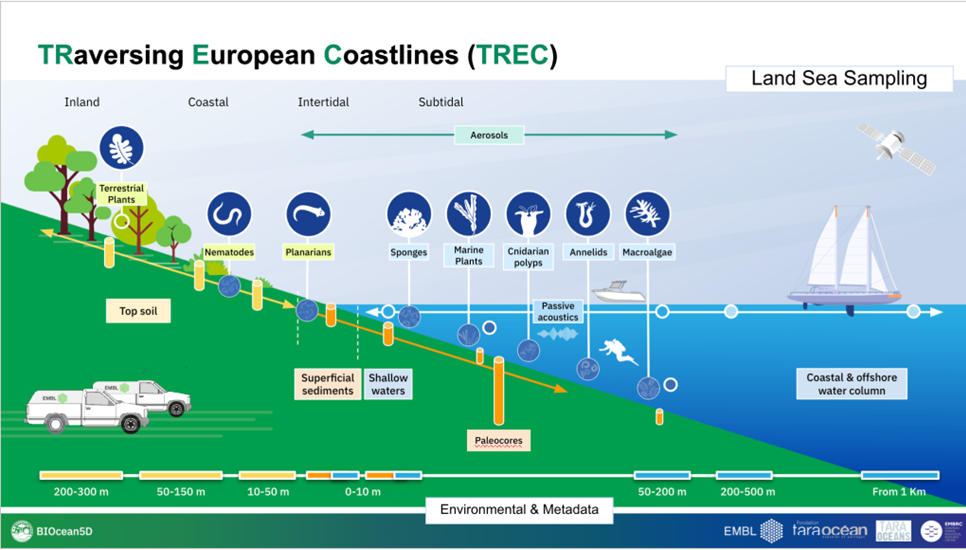
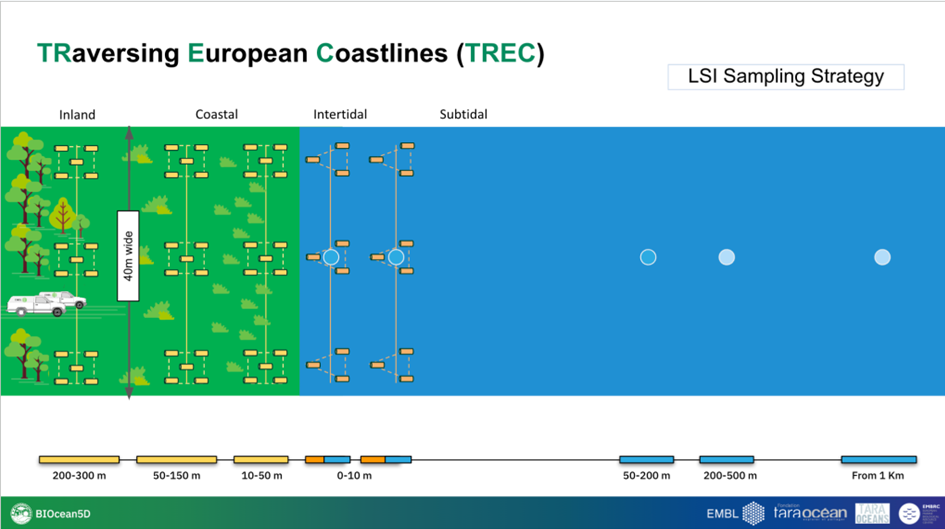
At each of the 120 sampling sites, we will take soil along 40 metres transects (parallel to the coast) at three different distances from the water and one or 2 sediment transects, done at intertidal and/or subtidal zones. Along each soil transect we will define 3 sampling areas of 1 metre per 1 metre and we will collect 5 soil subsamples per sampling area, producing a total of 3 soil samples (pool of 5 sub-samples per sample) in each soil transects. We will perform the same strategy for superficial sediments with the exception that each sampling area will contain only 3 subsamples, making 3 samples (pool of 3 sub-samples) per sediment transect. All samples will then be subsampled for molecular, physical, biogeochemical analysis and chemical profiling.
Analysis will include:
- on site probes measurements (such as temperature, pH, salinity, back-scatter, dioxygen)
- fluorescent dissolved organic matter
- Granulometry
- Porosity
- nutrients (nitrate, ammonium, phosphate)
- dissolved inorganic carbon
- organic carbon
- photosynthetic pigments
- metabolomic characterisation of dissolved and particulate organic matter
- metals
- organic pollutants
- environmental metagenomics
- environmental metatranscriptomics
- environmental metabarcoding (16S, 18S, ITS)
- Environmental metabolomics (including targeted and untargeted chemical profiling with focus on pesticide degradation products, antibiotics, synthetic hormones, TFAs, OPEs)
Quantities of soil and sediment samples
For sampling topsoil, we will use corers that are 20 cm deep with a 7 cm diameter. With a single core, we are able to take ~100-150g of soil, depending on the soil type. An individual sample is made up by pooling 5 homogenised cores, which makes around ~500-750g (5*100-150g) per soil sample. We take 3 samples (pooled of subsamples) at three distances from the coast, so this totals the quantity of soil taken at each sampling site between 1500 and 2250g, depending on the soil type and humidity conditions. From topsoil samples, nematodes will also be extracted for genomics and imaging analysis.
For sampling superficial sediments, we will use corers that are 10 cm deep x 15 cm and we will only collect the upper 5 cm. For sediment, one sample consists of 3 subsamples pooled, so we aim to have approximately 1590 ml per each of the three samples per sediment transect. We will take 3 sediment samples, so the total quantity of the sediment sample is around 4770 ml. From superficial sediment samples, meiofauna including nematodes will also be extracted for genomics and imaging analysis.
Where sampling will be performed
Soil and sediment samples will be collected at each sampling site from each sampling location. GPS locations are shown below in 4.1 Sampling locations in this document.
2.1.2 Sampling of shallow waters
To understand the land sea gradient as a continuum, the study of the interactions between the land and the sea are of main interest. Land and sea affect each other constantly: the sea splashes on the land and the soils affect the ocean by “washing out” the coasts with tides, by river inputs, underground waters and connected also by aerosols.
During TREC we will collect shallow waters that are defined as the waters of maximum 1 m depth, accessible directly from the shore. Shallow water sampling will happen along the 120 sites defined by the TREC land-sea transects and will be done directly from the shore and by foot. Tides will be taken into account.
Methods of shallow water sample collection
Shallow waters are collected in one spot that falls in the 40 metres sediment transect. The sampling is performed by walking into the waters until a depth between 30 and 100 cm deep. At the selected spot, the samplers immerse Niskin bottles that they bring back to the shore. The waters are accumulated in water containers on the shore until the team collects ~30-50L. 20 litres of the collected shallow waters are then filtered on a sampling filtration van (normally parked in the closest parking lot), size fractions are sorted and samples are then preserved. The rest of the collected waters are filtered once the sampling team is back at the local partner institute.
What will be analysed:
- on site probes measurements (such as temperature, pH, salinity)
- nutrients (nitrate, ammonium, phosphate)
- dissolved inorganic carbon
- organic carbon
- photosynthetic pigments
- metabolomic characterisation of dissolved and particulate organic matter
- metals
- organic pollutants
- environmental metagenomics
- environmental metatranscriptomics
- environmental metabarcoding (16S, 18S, ITS)
- Environmental metabolomics (including targeted and untargeted chemical profiling with focus on pesticide degradation products, antibiotics, synthetic hormones, TFAs, OPEs)
- Single cell ecology
Quantities of shallow waters sampling
For all genomics, we will collect approximately 20L of water which then will get filtered. In general, we will have 6 filters/site, the diameter of the filter is 142mm.
For chemical profiling, we will also collect approximately 20L. Still in finalisation but most probably the water will get filtered through 4*142mm filters per site, and 8 SPE columns per site.
For all other analyses (core parameters, single cell ecology), we will collect an additional 10L that will be fixed in PFA for single cell genomics.
Where sampling will be performed
Shallow waters will be sampled from each sampling location where soil and sediments are sampled.
2.1.3 Bioaerosols
Airborne microbes are known to impact the health of humans, plants, and animals. Additionally, they can drive cloud formation and affect key climatological processes. TREC intends to study the diversity, function and dispersal of airborne microbial communities at the land-ocean interface. Collected samples will be used to generate physico-chemical and genomic data including:
- environmental metagenomics
- environmental metabarcoding
We plan to sample aerosols in parallel with each soil sampling, at all locations.
Methods of sample collection
Sampling will be in parallel with the land-sea interface sampling of top soil, sediment and shallow water sampling, by setting up sampling equipment in the near proximity of the soil sampling locations.
Equipment used:
- 3x SASS 3100 Electret filter air sampler and 3 camera tripods (15 cm W x 17 cm L x 20 cm H and weight is 1.8 kg, noise levels are at 45–61 dB)
- EDM264 from GRIMM Aerosol Technik Ainring GmbH & Co. KG. The EDM264 instrument performs real-time monitoring of particle number and particle size and provides information on dust mass distribution. It will be running in parallel to the filter measurements (73 x 51 x 23 cm and weighs approximately 15 kg).
At each sampling location, 3 samplers will be deployed and running for 4 hours, producing 6 filters in total.
At selected sites, more extensive sampling will take place by deploying an additional sampler running for 5 days, producing 8 samples/day, generating 40 samples total.
All filters will be frozen in Whirl-Paks in liquid nitrogen and stored in a dry shipper. Samples will be shipped to EMBL to be added to the TREC sampling hub until redistributed to collaborators.
2.1.4 Sediment core sampling
Complementing the study of the present time with shallow sediments, TREC will use a paleogenomic approach (ancient DNA analyses) to study the microbial communities and species dynamics shifts in relation to punctual and extreme pollution activity related to human history (e.g. World War II) and the emergence of new microbes across the Anthropocene, and in particular in relation to the human industrial revolution started during the XIX century.
Methods of sample collection
For subtidal sediment cores, we will use a corer that is 1m x 9cm size. For collected sediment cores, we will use a boat with a winch to deploy the sediment corer (ideally owned by our local collaborators) and will collect the samples together with our local collaborators. 5 sediment cores will be taken.
Cores will be split in 1-cm layers, each of which will be subsampled for multidisciplinary analyses. Sediment subsamples will be collected for dating, granulometry, pollution and organic carbon, in order to characterise past environmental conditions and the evolution of pollutant concentrations across time. Sedimentary ancient DNA will be collected in each dated layer using all precautions needed for this parameter and in order to avoid contamination with modern DNA. Ancient DNA will be used for biodiversity analyses looking for i) community and species dynamics shifts in relation to punctual and extreme pollution activity related to human history (e.g. World War II) and ii) to chronic pollution developed during the industrialization time (industrial and agricultural contaminants). In addition, the multidecadal dynamics of potential invasive species will be reconstructed across Europe and linked to potential anthropogenic vectors of biological transports and to global seawater warming in the case of subtropical/tropical allochthonous species.
From the sediment cores, resurrection ecology of phytoplankton will also be studied, by resurrecting pre-Anthropocene populations of phytoplankton from sedimented resting stages.
What will be analysed:
Sediment subsamples will be collected for:
- Dating
- granulometry
- pollution
- organic carbon
- environmental metagenomics
- environmental metatranscriptomics
- environmental metabarcoding (16S, 18S, ITS)
Where sampling will be performed
For exact sampling locations, please refer to the GPS locations shown in 4.1 Sampling locations of this document.
2.2 Selected species and the environment
The selected species and phyla listed below will be sampled jointly, mostly by snorkelling or diving carried out by local scientific divers and with the use of their boats. In some cases the selected species will be hand-picked. The sampling strategy for all sampling activities was designed to not harm or disrupt the sampled habitats.
2.2.1 Platynereis dumerilii
The TREC project will sample the marine nereid Platynereis dumerilii from subtidal regions (atoke worms) and from the water column (epitoke worms), to investigate P. dumerilii diversity and its molecular and cellular adaptation to the environment. This will provide clues to (i) the population structure of P. dumerilii, (ii) the relevant microbiome, (iii) how different cells adapt to varying conditions, (iv) the correlation of the genetic standing variation and single-cell gene expression with morphological variation, to identify hotspots of cellular adaptation to environmental change and (v) how sensory organs adapt to environmental cues. Other polychaete and nemertean species will be collected to identify those who cohabit with Platynereis dumerilii.
Methods of sample collection
Immature worms: atokes will be collected by local divers who will retrieve a small amount of macroalgae or sea grasses (around 1kg) at 1-5 m depth. The divers will bring the samples to the surface and transport them to the local laboratory for worm sorting and preservation. Twenty atoke worms will be taken from each habitat. 5 individuals of each co-habiting polychaete and nemerteans species will be taken.
Co-occurring eukaryotes during atokes sampling:
When sorting macroalgae in buckets to find atokes Platynereis dumerilii, the water becomes muddy with macroalgae-associated grunge. That grunge is going to be mostly diatoms, microalgae, bacteria, and protists. The grunge will be filtered onto filter papers and will be snap freezed.
Epitoke worms: sexually mature Platynereis dumerilii worms will be collected during the evening from coastal superficial waters. Epitoke worms are sampled from a boat (ideally owned by our local collaborators). During the sampling, the sampling team attracts the mature P. dumerilii to the surface by using a light that attracts the worms, and collects the worms from the water surface with nets, without diving/snorkelling. Forty epitokes from the water column will be taken to spawn in the lab to generate larvae batches.
Analysis will include:
- Low coverage whole genome sequencing of individual worms
- Metatranscriptomics of individual worms
- Microbiome sequencing of individual worms
- Single-cell RNAseq of dissected worm heads
- Single-cell RNAseq of stage specific larvae
- Lipidomics of individual worms
- Metabarcoding of cohabiting polychaete and nemerteans
- X-rays imaging for phenotypic analysis of 6 days larvae
Where sampling will be performed
Depending on availability, Platynereis dumerilii will be sampled at each land-sea interface sampling site (soil, sediments, shallow waters), and also in the surrounding of our local collaborator’s marine station.
2.2.2 Seagrass (Zostera marina, Posidonia oceanica)
Seagrasses (host) and root symbiont clonal and genetic diversity will be addressed to better understand i) biogeography ii) host-microbe interactions in these associations and iii) the way the diversities of the holobionts respond to and are influenced.
Methods and quantities of sample collection
Collection is planned by snorkelling from a depth of 1-5 m, in parallel with the collection of Platynereis dumerilii and the deployment of the acoustic recording equipment, reaching the sampling points by local boats. The sampling does not harm the habitat and has no impact on the ecosystem function.
25 individuals (both the leaf and the roots) will be collected per sampling site. Additionally, seawater of 3×2 litres per location will be collected and filtered for microbiome analysis of the surrounding waters.
Analysis will include:
- Whole genome sequencing
- Transcriptomics (established 16S rRNA amplicon sequencing approaches combined with novel, primer-free direct long-read 16S rRNA sequencing for much improved resolution)
Where sampling will be performed
Seagrasses will be sampled at the same locations and in parallel with Platynereis dumerilii.
2.2.3 Lucinid clams (Loripes orbiculatus, Loripinus fragilis, Lucinella divaricata)
The use of seascape genomics, root/rhizome metagenomics, and associated lucinid bivalves in a continental-scale sampling across climates and different human impacts will reveal the dynamics of this three-player symbiosis and the influence it has on fragile seagrass ecosystems under different environmental factors. Little is known on the detailed mechanism and symbiotic evolution of the species of Sedimenticolaceae and the vital functions they provide for this ecosystem. To overcome this lack of knowledge, we are investigating the functions of the symbiont Sedimenticolaceae and the rest of the microbiome in seagrass root and rhizome and in symbiont Lucinid bivalves at more than 26 sites along the European coastline using metagenomics.
Methods and quantities of sample collection
At the same site (seagrass bed) selected for seagrass sampling, sediment will be collected from the surrounding of the seagrass bed. The sediment is collected at 20-30 cm depth. The ideal number of collected lucinid clams is around 30 individuals per sampling site, per species.
Analysis will include:
- Nucleic acid extraction
- metagenomic sequencing
- metatranscriptomic sequencing
- fluorescence microscopic imaging of bacterial symbionts
2.2.4 Sponges
TREC aims to study the effects of environmental factors and host genetics on the diversity and functionality of macro and microorganisms living on coastal regions. Many sponges harbour dense and diverse microbial consortia within their mesohyl tissue that collectively contribute to the fitness of the host sponge.
The effects of environmental change and extreme events on the structure and functioning of sponges and associated microbiomes will be investigated with metabarcoding, metagenomics and metatranscriptomics.
This study will establish (1) whether and how taxonomic composition and functional profiles of sponge holobionts change across temperature gradients, (2) whether microbial composition changes more than functioning or vice versa, and (3) how climatic extremes alter holobiont structure and functioning, potentially triggering mass-mortality events.
TREC will:
- seek to understand the effects of environmental gradients on variation and individuality
- visually document the morphotype, size, colour, and health/disease state of selected individuals
- will explore taxonomic and functional variation on the microbial symbiont side by barcoding and meta-omics, single cell genomics and microscopy
- will explore variation on the host side by population genomics and functionally, by exploring immune gene repertoires
- will provide high resolution insights into sponge-symbiont co-speciation and phylosymbioses.
Methods and quantities of sample collection
We will sample 4 sponge host holobionts (Agelas oroides, Chondrosia reniformis, Dysidea avara, Halichondria panicea). Sampling will be done by snorkelling or diving (either from the coast or from a boat, using local boats and divers), depending on depth. Smaller sponge tissue pieces will be cut from an intact individual sponge with a disposable scalpel, inflicting minimal damage to the organisms, or individuals will be scraped off from the substrate with a spatula getting the entire sponge needed. Once a sponge sample is obtained under water, it can be stored in a container (filled in the field with sea water beforehand) of different sizes to accommodate the sponge sample.
15 sponge individuals (tissue from 15 individual colonies) will be collected per sampling site; only a small part of the individuals will be sampled (approximately 5x5x5cm per colony). Therefore, the sponge will remain unharmed. Additionally, seawater of 3×2 litres per location will be collected and filtered for microbiome analysis of the surrounding waters.
Analysis will include:
- morphotype, size, colour, and health/disease state of selected individuals
- Barcoding and meta-omics
- single cell genomics
- Microscopy
Where sampling will be performed
Depending on availability, sponges will be sampled at each sampling location where seagrass are collected.
2.2.5 Leaves of terrestrial plants
Determination of the environmental features that lead to the increase or loss of leaf-associated microbial diversity and the metabolic pathways and responses involved will be addressed by analysing terrestrial plants biodiversity, microbiome and metabolomics status.
Methods and quantities of sample collection
A maximum of 8-10 leaves of 5 different non-protected terrestrial plants species will be collected by hand-picking, species depending on availability. Plan to collect samples at each sampling location. The leaves will be placed into separate paper envelopes and dried for until dried leaves crumble and show no signs of moisture. Dried leaves in paper bags will be placed air-tight in plastic bag(s) and prepared for shipping.
Analysis will include:
- DNA-based sequencing methods
- Masspectometry analysis
2.2.6 Planarians
Mechanisms of environmental adaptation in planarian flatworms will be studied by a molecular, cellular and physiological characterization of the animals in their natural habitats and in-depth functional study in the laboratory.
Methods and quantities of sample collection
Submerged rocks will be lifted, animals externally resembling planarian flatworms will be transferred to suitable accommodation using a paintbrush. Rocks will be returned to their original position so that each locality is left precisely as originally found.
Around 20 animals will be collected for genomic analysis. Around 40 animals will be collected per sampling site and preserved fixed, frozen or live to be analysed at EMBL and/or partner institutions.
Analysis will include:
- DNA and RNA sequencing
- Single cells multi-omics
- Histological characterization
- Functional studies in the laboratory using shipped live animals
Where sampling will be performed
Depending on availability, planarians will be collected around the coastal regions of the selected land-sea sampling sites (soil and sediment sampling sites) and in the local marine research institutes’ proximity.
2.2.7 Brown algae
Links among host-microbe genomic interactions and ecological success will be assessed in different species of brown algae.
Methods and quantities of sample collection
Samples will be taken by divers, in parallel with other sampling requiring diving. 30 individuals will be collected within a 15m transect with two individuals taken per metre.
Analysis will include:
- metabarcoding of swabs of the algal biofilm and surrounding sea waters for microbiome (including virome) analysis
- genome sequencing of the for host (vegetative blade tissue)
2.2.8 Live species for culturing: Fungi, marine (non-metazoan) Holozoans, Alveolates, phytoplankton, bacteria, viruses
Marine samples of fungi, marine (non-metazoan) Holozoans, Alveolates, phytoplankton, bacteria, viruses will be taken for subsequent culturing of these organisms. Sampling will not harm or disrupt the ecosystem. The cultures will be used for studying the cellular ultrastructure and developmental transitions in different organisms collected from sites with different environmental conditions.
Methods and quantities of sample collection
Plankton and protists smaller than 200 um will be collected using plankton net tows and sieving (2 um to 200 um sieves) from the specified coastal locations (relying on local boats). The samples will be concentrated and filtered using a manual pump. The concentrated sample will then be grown on both liquid cultures (single-cell sorted in 96-well plates, or directly in culture flasks; 30 culture flasks and 20 96-well plates) and solid medium (Petri- dishes with adequate antibacterial antibiotics, 40 petri dishes per super-site) to enrich for protists of interest that include unicellular holozoa, fungi, dinoflagellates, stramenopiles and ciliates. Pure cultures will be obtained by serial dilution, followed by species identification by 18s barcoding. The cultures will be used for studying cellular ultrastructure and developmental transitions using ultrastructure expansion microscopy. The sampling will not harm or disrupt the ecosystem.
The followings will be collected:
- 60 solid media plates (100 mm diameter)
- 1 litre sea water in culture flasks (~10 ml volumes, 20 cm2 flasks)
- Around 20 phytoplankton flasks per sampling site
- Around 10 glycerol stocks and 3 glycerol stocks
- Around 10 viral isolates
Analysis will mainly consist of:
- Ultrastructure expansion microscopy
- 18s barcoding for species identification
Where sampling will be performed
Collection will be done in parallel and at the same locations as plankton collection.
2.2.9 Lichens (species Trebouxia and Physcia)
Using 3D electron microscopy, TREC aims to understand the ultrastructural integration of the microalga within the fungi in lichen symbioses, as well as the morphological organization of the chloroplast.
Methods and quantities of sample collection
Sampling will be done from the shore by hand-picking. Few milligrams of lichens will be sampled from the rocks on the shore.
Samples will be cryofixed, for 3D electron microscopy (resin embedding) and for DNA barcoding to identify the different partners. Isolation and culturing of the microalgae from the lichens is also planned.
2.3 Assessing the biogeography of bioacoustic diversity – acoustic recorders
Coastal environments are ecological and anthropogenic hotspots. By applying ecoacoustic principles and methods, we will create acoustic maps based on the TREC sampling stations of the biophony (diversity of biological sounds from invertebrates to marine mammals) and the anthropophony (anthropogenic noise: e.g. noise budgets). This non-invasive sampling will occur using underwater passive acoustic recorders, moored for at least 48h. This will allow a punctual assessment of the acoustic biodiversity of a site, the identification of key species (sentinel species, structuring species, etc) and functionally relevant sites (such as reproduction sites); and to characterise noise pollution. This can lead to state-pressure indices of marine environments. The acoustic data will also be linked to other sampled data such as eDNA, to assess potential relationships between sound presence and species presence; and help establish the sources of acoustic biodiversity, identify species or functional groups related to soundscape features, that may be relevant for ecosystem functioning & condition.
Methods of sample collection
Underwater passive acoustic recorders will be deployed, moored for 24-48 hours, and collected by local divers, from a local boat (ideally owned by our local collaborators). Deployment is in parallel with other sampling activities from a boat, e.g. Platynereis dumerilii and seagrass sampling.
Where sampling will be performed
Passive acoustics recorders will be deployed in parallel and at the same locations as Platynereis dumerilii and seagrasses are collected.
2.4 Planktonic uni- and multi- cellular organisms
Advanced imaging is key to visualise microplankton’s morphology and ultrastructure. During TREC, a large spectrum of microscopy modalities will be utilised to identify species (taxonomy) and to explore plankton biodiversity at an unprecedented resolution, in combination with advanced sequencing approaches. Subcellular imaging will reveal if and how geographical or environmental gradients affect their morphologies across spatial scales.
Methods and quantities of sample collection
Plankton will be collected on a daily basis using boats provided by the host marine station or rented from private providers. On board, samplers deploy plankton nets of various mesh sizes (from 5 to 150 µm) that are towed for 2 to 10 minutes. The nets are immersed at specific depths as determined by measurement of CTD profiles (especially aiming for the chlorophyll peak). Each net tow yields c.a. one litre of sea water containing enriched planktonic fraction that is poured through sieves of various mesh sizes to establish plankton size fractions: 5-20µm; 10-40µm; 20-200µm; 150-400µm. The fractions are then brought back to land where they are processed inside the Advanced Mobile Laboratory.
Where sampling will be performed
Plankton collections will be collected around the coastal regions of the selected land-sea sampling sites (soil and sediment sampling sites) and in the local marine research institutes’ proximity.